Understanding points
Understanding points
D1.2.1 Transcription as the synthesis of RNA using a DNA template
D1.2.2 Role of hydrogen bonding and complementary base pairing in transcription
D1.2.3 Stability of DNA templates
D1.2.4 Transcription as a process required for the expression of genes
D1.2.5 Translation as the synthesis of polypeptides from mRNA
D1.2.6 Roles of mRNA, ribosomes and tRNA in translation
D1.2.7 Complementary base pairing between tRNA and mRNA
D1.2.8 Features of the genetic code
D1.2.9 Using the genetic code expressed as a table of mRNA codons
D1.2.10 Stepwise movement of the ribosome along mRNA and linkage of amino acids by peptide bonding to the growing polypeptide chain
D1.2.11 Mutations that change protein structure
D1.2.12 Directionality of transcription and translation (HL only)
D1.2.13 Initiation of transcription at the promoter (HL only)
D1.2.14 Non-coding sequences in DNA do not code for polypeptides (HL only)
D1.2.15 Post-transcriptional modification in eukaryotic cells (HL only)
D1.2.16 Alternative splicing of exons to produce variants of a protein from a single gene (HL only)
D1.2.17 Initiation of translation (HL only)
D1.2.18 Modification of polypeptides into their functional state (HL only)
D1.2.19 Recycling of amino acids by proteasomes (HL only) |
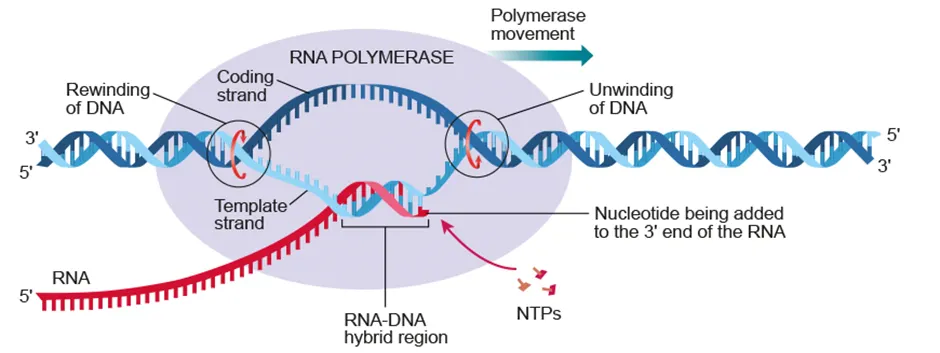
Transcription
•
Production of mRNA from DNA by RNA polymerase
RNA polymerase unwinds DNA and binds to the promoter with the help of transcription factors
↓
*(AHL) RNA is synthesized in 5’ → 3’ direction by complementary base pairing of A-U and G-C
↓
RNA and RNA polymerase detach from template + DNA rewinds into double helix |
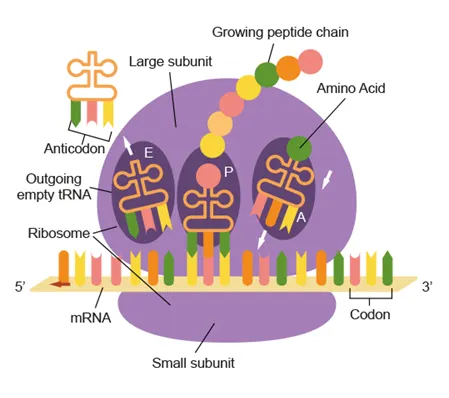
Translation
•
Production of a polypeptide from mRNA by ribosomes
•
Ribosomes are made of rRNA and protein
◦
Large subunit: 3 binding sites (A,P E) for tRNA
◦
Small subunit: mRNA binding site
•
Codon: 3 bases on mRNA that code for an amino acid
◦
Complementary base pairing with anticodon on tRNA
*(AHL) Translation initiates at start codon (AUG)
↓
mRNA binds to ribosome small subunit
↓
tRNA binds with αα then attaches to ribosome (1st tRNA at P site)
↓
Anticodon on tRNA pairs with codon on mRNA
↓
Peptide bond forms between the amino acids of consecutive tRNAs
↓
The 1st tRNA leaves via the E site and the 2nd tRNA enters the A site
↓
As ribosome moves along the mRNA, polypeptide elongation occurs
↓
Translation stops when it reaches a stop codon (UAA, UAG, UGA) |
Sickle cell anemia
Cause | Base substitution mutation in hemoglobin gene
Single base change from CTC to CAC
𝛼𝛼 change from glutamic acid to valine |
Result | Hemoglobin forms fibrous, insoluble strands
RBC becomes sickle shaped and cannot carry sufficient oxygen |
Genetics | Sickle-cell allele is codominant
Heterozygous = malaria resistant carrier
Homozygous = anemia |
*(AHL)
Post-transcriptional modification
•
In the nucleus
Removal of introns | Introns are removed and exons are spliced together
Alternative splicing: different exons joined to produce different polypeptides |
Addition of 5’ cap | Methylguanine is added to 5’ end of mRNA |
Addition of 3’ tail | 100~200 adenine nucleotides are added to 3’ end of mRNA |
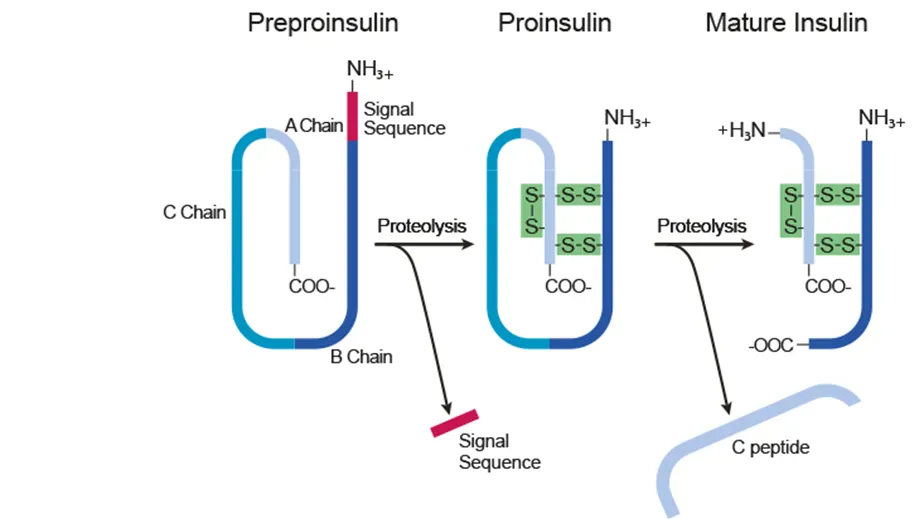
Post-translational modification
Noncoding sequences
Gene regulators | Promoters and repressors |
Introns | Removed from mRNA before translation |
Telomeres | Repetitive sequences at the ends of chromosomes that protects DNA |
tRNA and rRNA genes | Transcripted, but not translated |
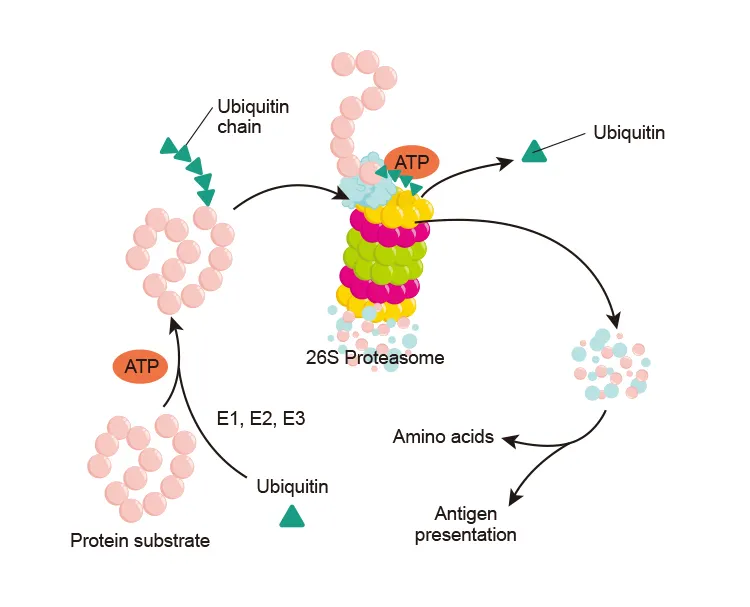
Proteasomes
•
Degrade damaged proteins marked with ubiquitin